Thesis prepared by Philippe Gantzer: "Development and comparison of inverse QSPR-inverse approaches"
What chemical engineer has never dreamed of having access to a tool that can directly identify a fluid (pure substance or mixture) on the basis of characteristics necessary to a given application context? This Holy Grail could become a reality thanks to the field of Chemoinformatics and its methods.
Cheminformatics lies at the interface of several scientific fields and involves using computer and information science techniques to solve problems related to chemistry. One application involves using artificial intelligence to predict usage properties on the basis of reference data, relative to composition or structure [1]. Using machine learning, it is thus possible to establish models that are bridges between molecular descriptors and properties of interest. In addition to this predictive use, chemoinformatics can be used in screening processesa, or even be part of an inverse approach, i.e., to propose molecular structures likely to satisfy certain constraints.
With this in mind, the specific aim of this PhD research was to develop a molecular structure generation method. Having identified the different methods in the literature [2], two approaches were selected: the concatenation of molecular fragments (CFM) and the evolution of molecular structures (ESM). The first consists in identifying molecular fragments and then combining them, while the second consists in changing molecular structures via the application of different operators, such as atom hybrids or mutations, bonds or functional groups. Once implemented in numerical form, these approaches were compared with one another using a specific approach.
There are, in fact, numerous criteria for evaluating the predictive quality of a model describing properties, but very few existed for new structure generation methods. To overcome this, we proposed a specific approach as illustrated in the figure below. This consists in projecting virtual molecular structures in the “chemical space” – a multidimensional space based on molecular descriptors -b (and reduced to three dimensions here) – and then comparing these projections on the basis of indices reflecting their degree of occupationc or coveraged of this space [3].
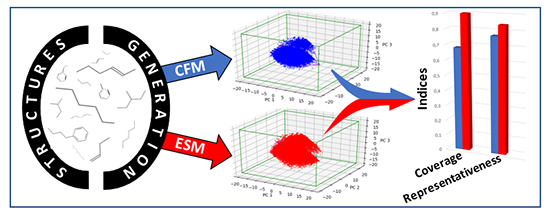
The comparisons made on the basis of these new indices showed that the ESM approach is more effective than the CFM method. If offers better coverage of the chemical space and is thus capable of proposing a greater diversity of new structures meeting given specifications.
Therefore, it is this method that is going to be used in different application contexts, such as to identify new solvents.
a- Identification of molecules in an existing base.
b- Such as a breakdown of functional groups.
c- Quantity of elementary cubes occupied.
d- Individual population of elementary cubes.
Bibliographic references:
-
Issue 48 of Science@ifpen, June 2022.
>> https://www.ifpenergiesnouvelles.com/brief/cheminformatics-and-its-descriptors-application-polymerfluid-compatibility
-
Gantzer, P.; Creton, B.; Nieto-Draghi C. "Inverse-QSPR for de novo Design: A Review", Molecular Informatics 2020, 39(4), 1900087.
>> https://doi.org/10.1002/minf.201900087
-
Gantzer, P.; Creton, B.; Nieto-Draghi C. "Comparisons of Molecular Structure Generation Methods Based on Fragment Assemblies and Genetic Graphs", Journal of Chemical Information and Modeling 2021, 61(9), 4245-4258.
>> https://doi.org/10.1021/acs.jcim.1c00803
Scientific contacts: benoit.creton@ifpen.fr ; carlos.nieto@ifpen.fr
You may also be interested in
Microfluidics and Chemoinformatics: a highly complementary approach to studying material/fluid compatibility
Pour de nombreuses applications industrielles, comme le recyclage chimique des plastiques, ou encore pour assurer la compatibilité entre polymères et nouveaux carburants, il est essentiel d’anticiper les interactions entre matériaux et fluides...
Cheminformatics and its descriptors: application to polymer/fluid compatibility
Ensuring compatibility between polymers and fluids is essential in numerous industrial sectors: in the automotive sector, for example, the resistance of materials used in the fuel supply system is a vital consideration.