18.01.2019
5 minutes of reading
IFPEN’s Scientific Board awarded the 2018 Yves Chauvin prize to Aurélie Pirayre for her thesis entitled “Reconstruction and Clustering with Graph optimization and Priors on Gene networks and Images”. Aurélie Pirayre received her award at the ceremony held at IFPEN’s Rueil-Malmaison site on 19 November 2018.
Her work represents an advance in the adaptation of image processing methods for the purposes of representing biological data in graph form. Directed by Jean-Christophe Pesquet (Université Paris-Est Marne-la-Vallée, now CentraleSupélec), her research was supervised by Frédérique Bidard-Michelot and Laurent Duval at IFPEN.
After having defended her thesis in July 2017, Aurélie Pirayre joined IFPEN’s Digital Sciences and Technologies Division.
What was the focus of your research?
The aim of my research was to improve our understanding of the mechanisms involved in the production of enzymes by the fungus, Trichoderma reesei. These specific enzymes are used as biocatalysts to degrade biomass into simple sugars in certain second-generation bioethanol production processes. My work consisted in developing mathematical approaches to elucidate the genetic functioning of this fungus, in order to be able to improve the enzyme production process.
The research was driven in collaboration between IFPEN’s “Biotechnology” and “Control, Signal and Systems” departments in order to take advantage of their expertise in two fields: biology and mathematics for image processing and optimization. In addition to initiating the bioinformatics at IFPEN, the developed approach led to new tools allowing a better exploitation and analysis of biological data.
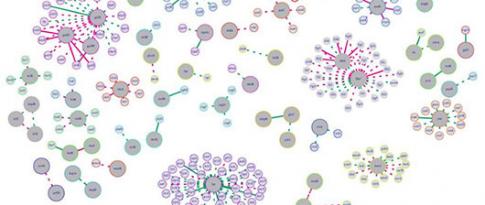
As a result, we were able to show, in graph form, the reaction cascades between genes describing the genetic functioning of Trichoderma reesei. The nodes of the graph correspond to genes and the links between the nodes correspond to the interactions between the genes. To obtain biologically realistic networks, we integrated biological information into adapted image processing methods. That is what is original about my research: we took biological knowledge and translated it into mathematical formulae in order to incorporate it in existing image processing models.
Using this approach, we developed several tools, grouped together under a generic name: BRANE (Biologically-Related Apriori for Network Enhancement). Two main tools - BRANE Cut and BRANE Clust - were published in leading scientific journals in the field of bioinformatics and the research was also published in specialized biology and mathematics journals.
Will the research be continued?
The Trichoderma reesei fungus is still a relatively unknown quantity. My research has elucidated some complex regulating mechanisms for the production of biocatalysts.
The research will continue in the context of two theses that will examine the same basic problem, i.e. improving our understanding of the genetic functioning of T. reesei. One thesis will concentrate more on the mathematics, while the other will focus on the biology. Both these theses will be followed by the same team that for my thesis.
As for the BRANE software suite, the aim is now to extend its use to new projects concerning other microorganisms studied by IFPEN. The BRANE suite was developed for the purposes of being routinely used by biologists, with their data. Bioinformatics is a fast-developing area and new biological data acquisition technologies are being implemented, generating huge amounts of data. We encounter all the problems associated with what’s known as “Big Data”, with large volumes of heterogeneous data, and that’s when mathematics and IT become essential to biologists, so that they can process this data and extract useful information.
What did you appreciate about carrying out your research at IFPEN?
IFPEN is a great place to do a thesis. PhD researchers are very well looked after. A whole array of measures are in place to help them feel part of the team: integration days and themed seminars, for example. PhD researchers have access to a very high-quality scientific environment with lots of training opportunities, a real plus for those hoping to continue working in the field of energy and transport. There is also a PhD researchers association, the ADIFP, which offers activities such as English lessons or study trips, and which is actively involved in the organization of various events, including, for example, "Doc Day IFPEN”, when researchers have the opportunity to meet companies.
During my thesis, the supervising team was always accessible and attentive, particularly since this was the first time a bioinformatics theme had been studied at IFPEN. There were teams specializing in mathematics and biology, two completely different areas that do not use the same vocabulary. I was chosen for my dual expertise, allowing me to span these two fields relatively quickly and easily. There really is an atmosphere of mutual listening in order to accurately define problems and try to solve them together. This team spirit was crucial to the success of my research.
Publications
- A. Pirayre, C. Couprie, F. Bidard, L. Duval, J-C. Pesquet. BRANE Cut: biologically-related a priori network enhancement with graph cuts for gene regulatory network inference, BMC Bioinformatics, 2015.
>> DOI: 10.1186/s12859-015-0754-2
- A. Pirayre, C. Couprie, L. Duval, J-C. Pesquet. BRANE Clust: Cluster-Assisted Gene Regulatory Network Inference Refinement, IEEE/ACM Transactions on Computational Biology and Bioinformatics, 2018, Vol. 15, Issue 3.
>> DOI: 10.1109/TCBB.2017.2688355
You may also be interested in
‘‘BRANE Power’’: of genes and algorithms, an alliance for green chemistry
THESIS BY AURÉLIE PIRAYRE, 2018 YVES CHAUVIN PRIZE