23.04.2020
3 minutes of reading
To optimize the breakdown of biomass required for bioethanol production, a research thesis conducted at IFPEN focused on studying new enzymes derived from fungal strains belonging to the genus Aspergillus. The research identified a new family of enzymes able to increase cellulase activity.
For a more efficient Trichoderma reesei enzyme cocktail
Lignocellulosic biomass is an abundant and renewable alternative to fossil resources, but its efficient break-down is a major challenge for the development of a low-cost bioethanol process.
This challenge was the focus of a PhD thesis [1], conducted in partnership with INRAE’s “Biodiversité et Biotechnologie Fongiques” (BBF - Fungal Biodiversity and Biotechnology) laboratory in Marseille. The aim was to search for new enzymes able to increase the efficiency of the Trichoderma reesei enzyme cocktail, which is already used in second-generation ethanol production processes for the breakdown of biomass prior to fermentation.
Complementary enzymes in Aspergillus strains
In a previous ANR (E-TRICEL1) project, fungal strains belonging to the genus Aspergillus and producing enzymes that are complementary to T. reesei cellulases had been identified. Following on from that study, the “secretomes” (= the whole set of secreted enzymes) ) of 5 Aspergillus strains were studied for their capacity to improve the activity of the reference cocktail.
Two of these secretomes, produced by an Aspergillus japonicus strain, showed the capacity to significantly enhance the hydrolysis of a biomass of interest, Miscanthus grass. They contained a protein with structural similarities to LPMOs (lytic polysaccharide monooxygenases2), in particular the presence of conserved Histidine3 residues that typically coordinate a Cu2+ ion (diagram).
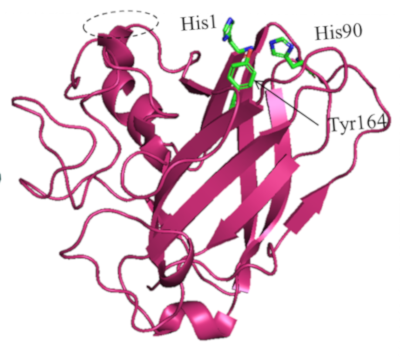
Enzyme tests then confirmed that this protein possessed an oxidative activity and was able to improve the activity of the T. reesei CBH1 cellulase on cellulosic substrates [2].
AA16: a new family to be characterized
Since these new enzymes have a structure that differs from those of known LPMOs, they represent a new family in the CAZy database (www.cazy.org) classifying all carbohydrate-active enzymes.
Subsequently, several other members of this new family, named AA16, demonstrated the same capacity to release oxidized sugars and increase cellulase activity.
In-depth characterization of AA16 enzymes will be the focus of another PhD thesis. In particular, the objective will be to determine the identity of optimal redox partners, as well as their three-dimensional structure, via crystallography, and to study the synergistic activity with cellulases on real lignocellulosic substrates.
1 Entitled Exploration de la biodiversité enzymatique pour la complémentation du secrétome de Trichoderma reesei afin d’améliorer l’hydrolyse des lignocelluloses (Exploration of enzyme biodiversity for complementation of the Trichoderma reesei secretome in order to improve lignocellulosic hydrolysis)
2 Enzymes that cleave polysaccharides via an oxidative mechanism
3 One of the amino acids constituting proteins
References :
[1] Camille Filiatrault-Chastel’s thesis: “Exploration de sécrétomes d’Aspergillus spp. en vue d’une complémentation du cocktail cellulolytique de Trichoderma reesei” (Exploration of Aspergillus spp secretomes, for the complementation of the Trichoderma reesei cellulolytic cocktail), 2019.
[2] Filiatrault-Chastel, C., Navarro, D., Haon, M., Grisel, S., Herpoël-Gimbert, I., Chevret, D., Fanuel, M., Henrissat, B., Heiss-Blanquet, S., Margeot, A., and Berrin, J.-G. AA16, a new lytic polysaccharide monooxygenase family identified in fungal secretomes. Biotechnol Biofuels 12, 55 (2019), DOI: 10.1186/s13068-019-1394-y
Scientific contact : senta.blanquet@ifpen.fr
> To find out more: Fermentation and Ethanol





