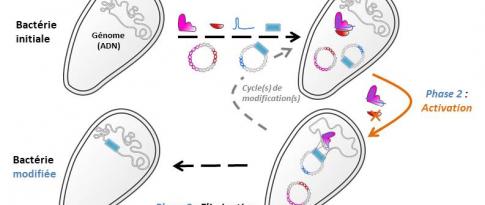
Most frequently, the use of a microorganism in the production of biofuels or biosourced chemicals requires its optimisation. This is achieved through genetic engineering, which involves inactivation and/or addition of one or more genes, to improve the ability of this microorganism to produce a target molecule. Identifying genetic engineering strategies requires a solid knowledge of the complex biological systems in which the improvement is needed.
The explosion of “omics” analysis techniques over the last twenty years has made it possible to identify and quantify the biological molecules that make up a cell in their totality and to gain a better understanding of their phenotype (the observable behaviours, e.g. the production of a molecule of interest). Now, multiple levels of data can be collected to establish the relationship between the various macromolecules (DNA, RNA, proteins, metabolites) of a biological system (figure).
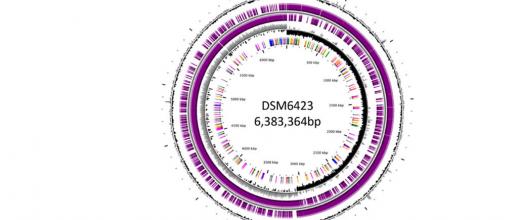
At IFPEN, access to genomics (DNA) has helped in gathering data on the composition and structure of the genomes of the microorganisms used in the Biotechnology division [1,2]. This means we can now accurately map the genetic code of the industrial strains produced, then link it to the phenotypes observed.
Click on the picture to enlarge
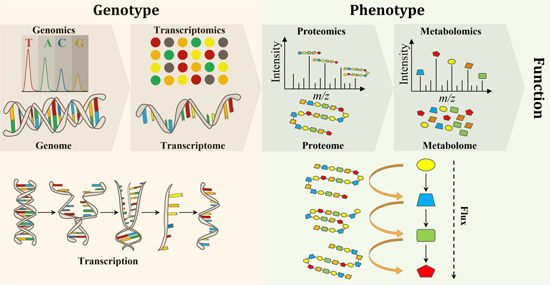
The following stages of analysis involve monitoring the expression of the thousands of genes of a micro-organism and the synthesis of their product, through transcriptomic (RNA), proteomic (proteins) and metabolomic (metabolites) approaches. The tools developed through these approaches [4,5] are now used as standard, including for research theses, in order to answer questions related to the optimisation of our microorganisms. They help to fine-tune our fermentation processes. The results achieved as part of a thesis by Rémi Hocq, who received three awards for his work in 2020-2021, are primarily based on the use of these tools.
More recently, methods of gathering information on the epigenetic regulationa of gene expression were developed as part of another doctoral paper. Epigenomic data have made it possible to identify previously unknown regulation mechanisms involved in the production of cellulases in Trichoderma reesei, a model microorganism involved in our lignocellulosic biomass hydrolysis procedures [6].
Finally, research into new enzymatic activities or new microorganisms is carried out through the analysis of complex ecosystems. In this context, metagenomic and/or metatranscriptomic approaches are deployed. These approaches allow us not only to establish the microbiological composition of a specific biotope - such as an environment that is likely to contain microorganisms of interest, or subsurfaces linked to renewable energy projects (geothermal or hydrogen storage) - but also to identify the enzymes present and the active synthesis routes.
In summary, the use and mastery of “omics” tools at IFPEN has a two-fold effect, giving us a more in-depth understanding of the microorganisms of interest and optimising them for use in breakthrough bioprocesses.
a- An area of biology that studies the nature of mechanisms that modify gene expression reversibly, transferrably and adaptively without altering the nucleotide sequence.
References:
- Jourdier, E., Baudry, L., Poggi-Parodi, D., Vicq, Y., Koszul, R., Margeot, A., Bidard, F. (2017). Proximity ligation scaffolding and comparison of two Trichoderma reesei strains genomes. Biotechnology for biofuels, 10 (1), 1-13.
>> doi.org/10.1186/s13068-017-0837-6
- Máté de Gérando, H. M., Wasels, F., Bisson, A., Clement, B., Bidard, F., Jourdier, E., … & Ferreira, N. L. (2018). Genome and transcriptome of the natural isopropanol producer Clostridium beijerinckii DSM6423. BMC genomics, 19 (1), 1-12.
>> doi.org/10.1186/s12864-018-4636-7
- Amer, B., & Baidoo, E. E. (2021). Omics-Driven Biotechnology for Industrial Applications. Frontiers in Bioengineering and Biotechnology, 9, 30.
>> doi.org/10.3389/fbioe.2021.613307
- Hocq, R., Jagtap, S., Boutard, M., Tolonen, A.C., Duval, L., Pirayre, A., Lopes Ferreira, N., & Wasels, F. Genome-wide TSS distribution in three related Clostridia with normalized Capp-Switch sequencing. Microbiology Spectrum, e02288-21.
>> doi.org/10.1128/spectrum.02288-21
- Poggi-Parodi, D., Bidard, F., Pirayre, A., Portnoy, T., Blugeon, C., Seiboth, B., … & Margeot, A. (2014). Kinetic transcriptome analysis reveals an essentially intact induction system in a cellulase hyper-producer Trichoderma reesei strain. Biotechnology for biofuels, 7 (1), 1-16.
>> doi.org/10.1186/s13068-014-0173-z
- Fajon S., Jourdier E., Bidard F. (2021). Souche de champignon hyperproductrice de protéines. Patent 21/11.274
Glossary
DNA: nucleic acid, carrier of genetic information
RNA: nucleic acid allowing the synthesis of proteins
Metabolite: intermediate organic substance or product of metabolism
Epigenetics: study of the nature of mechanisms that modify the expression of genes without altering their sequence
Metagenomics: the entire set of genes contained in a given biotope, all species combined
Metatranscriptome: the entire set of genes expressed in a given biotope, all species combined
Cellulase: biocatalyst capable of breaking down cellulose (biopolymer)
Scientific contacts: Frédérique Bidard-Michelot, Francois Wasels
You may also be interested in
Hydrolysis of lignocellulosic biomass: study of enzyme-substrate interactions (HDR 2015)
The scope of my HDR covered ten years of research at IFPEN within the context of the development of Futurol™, a process aimed at producing 2nd-generation bioethanol from lignocellu
‘‘BRANE Power’’: of genes and algorithms, an alliance for green chemistry
THESIS BY AURÉLIE PIRAYRE, 2018 YVES CHAUVIN PRIZE