14.05.2024
3 minutes of reading
“Lytic Polysaccharide MonoOxygenase“ (LPMO) enzymes are widespread enzymes with a copper atom in the active site. In the natural environment, they stimulate the conversion of biopolymers (for example, cellulose or chitin) [1]. For several years now, IFPEN and INRAE have been working on gaining a better understanding of these enzymes [2, 3] and also on the cellulases produced by the T. reesei fungus, with a view to their use for bio-based industrial processes.
Fungi possessing up to 60 coding genes for AA9 LPMOs
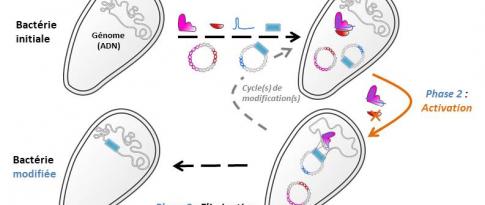
A PhD thesis [4] led by the two institutes and supported by the ANR SNOEBORD project studied a sub-family of LPMOs possessing atypical and previously unknown properties [5]. Some polypores, fungi that decay wood (white rot), can possess up to 60 coding genes1 for LPMOs belonging to the AA9 LPMO family2. However, the functional relevance of such redundancy has yet to be discovered. Previous comparative transcriptomic studies of six Polypore fungi cultivated on cellulosic substrates demonstrated the overexpression of numerous coding genes for AA9, including some with a C-terminal domain3 of unknown function “X282” (figure 1).
1 Gene whose translation leads to the formation of a protein
2 AA9: Enzymes classified as having an Auxiliary Activity (AA) family 9 according to reference nomenclature
3 Enzymes are amino acid chains linked by a peptide bond. The enzyme’s first amino acid thus possesses a free amino called N-terminal and the last a free carboxylic acid function, called C-terminal.
A sub-family of AA9 LPMO has been shown to have strange phosphate affinity
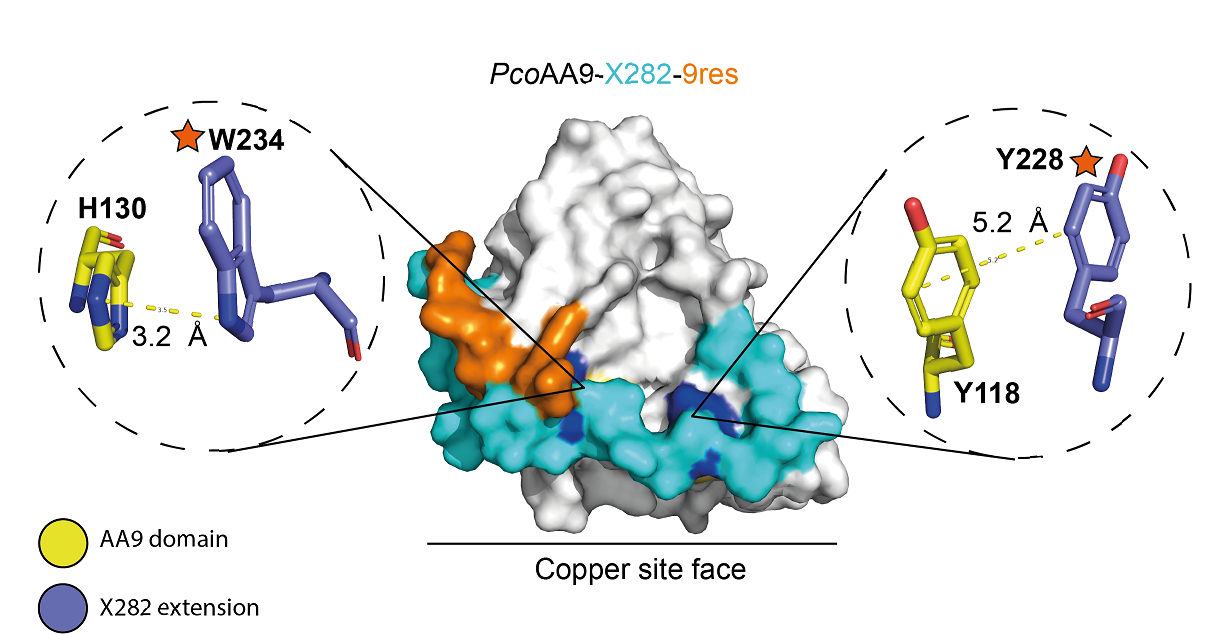
On the basis of structural predictions and phylogenetic analyses4, we selected and characterized six AA9-X282 LPMOs with different domains and atypical characteristics. Unexpectedly, having examined a broad range of conditions, these AA9-X282s only demonstrated weak cellulose bonding properties and little or no cellulolytic oxidative activity. The proteomic analysis then revealed the presence of multiple phosphorylated residues on the surface of these AA9-X282 LPMOs, including a conserved residue near the copper site. More in-depth analyses relating to a glycine-rich C-terminal extension with 9 residues suggested it may possess phosphate binding properties.
4 A phylogenetic analysis is used to compare the DNA sequence of genes from the same family
LPMO AA9 enzymes with more than one trick up their sleeve
The results of this work partially call into question the involvement of AA9 LPMO enzymes in the degradation of plant cell walls, and open up new avenues to explain the divergence in function of certain LPMO enzyme members of the AA9 fungal family.
References
[1] Vandhana, T.-M., Reyre, J.-L., Sushmaa, D., Berrin, J.-G., Bissaro, B., Madhuprakash, J., On the expansion of biological functions of lytic polysaccharide monooxygenases. New Phytol. 2022 Mar;233(6):2380-2396. DOI: https://doi.org/10.1111/nph.17921
[2] Thesis by Camille Filiatrault-Chastel : « Exploration de sécrétomes d’Aspergillus spp. en vue d’une complémentation du cocktail cellulolytique de Trichoderma reesei » (Exploration of Aspergillus spp secretomes, with a view to complementing the Trichoderma reesei cellulolytic cocktail), Paris, Institut agronomique, vétérinaire et forestier de France, 2019.
[3] Filiatrault-Chastel, C., Navarro, D., Haon, M., Grisel, S., Herpoël-Gimbert, I., Chevret, D., Fanuel, M., Henrissat, B., Heiss-Blanquet, S., Margeot, A., and Berrin, J.-G., AA16, a new lytic polysaccharide monooxygenase family identified in fungal secretomes. Biotechnol Biofuels 12, 55 (2019), DOI: https://doi.org/10.1186/s13068-019-1394-y
[4] Thesis by Jean-Lou Reyre : “ Lytic Polysaccharide Monooxygenases (LPMO)" Fongiques : Mise en œuvre en bioraffinerie et découverte de nouvelles enzymes aux propriétés atypiques» (Fungal Lytic Polysaccharide Monooxygenases (LPMO): Implementation in a biorefinery and discovery of new enzymes with atypical properties), Université Paris-Saclay, 2023
[5] Reyre, J.-L., Grisel, S., Haon, M., Xiang, R., Gaillard, J.-C., Armengaud, J., Guallar, V., Margeot, A., Arragain, S., Berrin, J.-G., Bissaro, B. Insights into peculiar fungal LPMO family members holding a short C-terminal sequence reminiscent of phosphate binding motifs. Scientific Reports 13, 11586 (2023), DOI: https://doi.org/10.1038/s41598-023-38617-5
Scientific contact : Simon Arragain
You may also be interested in
Hydrolysis of lignocellulosic biomass: study of enzyme-substrate interactions (HDR 2015)
The scope of my HDR covered ten years of research at IFPEN within the context of the development of Futurol™, a process aimed at producing 2nd-generation bioethanol from lignocellu
Extrapolating ethanol-from-biomass production technologies
The industrial production of ethanol from lignocellulosic biomass was demonstrated by the Futurol™ projecta.